Ena Dzanko
DATA ENGINEER
INTRODUCTION
The importance of our brain and cognitive abilities is something many of us don’t think twice about during our everyday activities. Nevertheless, none of our cognitive, normal functioning would be possible if it wasn’t for this complex structure we like to call our brain. So, why wouldn’t we devote a few words, (or a whole blog), to the research area of IT in neurology and neuroscience, so we can take a small step towards realizing the power of computer science and computer engineering, in an area we would consider ourselves helpless as data engineers.
Neuroscience is a collected set of multidisciplinary sciences that analyze the nervous system, where each discipline cooperates to understand structures and functions of a normal and abnormal brain. As it is a set of disciplines, we could introduce molecular, cellular, cognitive, behavioral, and other neurosciences, but will focus on the areas such as computational neuroscience and more importantly – neural engineering (neuroengineering), a discipline that uses engineering techniques to understand and impact neural systems. Neuroscience was first introduced in the 1960s and today is one of the fastest growing areas of science, with a wide range of approaches (clinical, cultural, cognitive, developmental, molecular, computational and other) which, for instance, provide good diagnostics methods (neuroimaging), life assistance options and even disease prediction. Despite that, we have still made relatively small progress in comparison to what we possibly could know by using computer engineering, mainly by manipulating large datasets and “extracting” significant information for not yet discovered solutions.
In the last few years of neuroscience development, we’ve seen and have usually connected the terms “neuro” and “science” in IT with pure data science, i.e., machine learning and deep neural networks. A minority, however, see the importance of data engineering as an inevitable step which precedes those famous data science workloads.
This blog will briefly cover the basic concept of neurology, brain structure and brain functioning needed for an engineer’s understanding of how this complex, nonlinear system works. We will then cover our data engineering perspective and its role in neuroscience, by considering and explaining neuroengineering use cases, data models, data formats, neuroethics and neuro-data governance.
But I am not a neuro-scientist?
By hearing the term “Neuroscience in IT” we would first think of a data scientist in the medical industry, who creates neural networks or machine learning models for certain brain insights or potential clinical assistance. A data scientist uses complex mathematical models for deeply analyzing (neuro)data and providing conclusions and metrics, therefore possible revolutionary solutions. But how can a data engineer help in this area? A data engineer comes in the previous step, where he is needed for the development of solutions for governance and useful application of that medical data and solution. A data engineer can focus on different work areas and techniques, ranging from medical data collection and processing, all the way to medical data management and application.
Into the brain
As engineers, we know to define neural networks as a series of algorithms with the goal of recognizing underlying relationships in data. But how much do we really know about how the real brain works? An average person would probably say it is a complex system of nerves, but hardly any more than that.
The brain’s structure and functions are mainly driven by highly specialized cells, called neurons, which are interconnected and bioelectrically driven. Each neuron will communicate with thousands of others through synapses by transferring electrical impulses, therefore, creating a highly dynamic and nonlinear environment. Everything that happens in the brain, including our thinking, memorizing, motor skills and everyday physical processes like breathing, chewing, and blinking, is a consequence of the impulses that are passed between connected neuron cells and are recognized as signals. Signals that we can convert and analyze as medical data.
Although the brain, in its full glory, is powerful, to this day it stands helpless against common neurocognitive diseases, like Schizophrenia, Epilepsy, Alzheimer’s, Parkinson’s, Dementia and many others. In most cases, neurologists can diagnose but not significantly help those patients. Although we have made decent progress in patient monitoring systems which can improve the quality of life in such patients, as of today we only have the medically discovered (not explained) causes of most of these diseases without a cure. These diseases, by affecting the brain, affect the quality of life for the individual, as well as their caregivers, since some are characterized as progressive – meaning irreversible and at this current point, incurable. They can manifest in different, yet similar ways. For example, many of them include some type of sleep disorder, psychological changes, and cognitive decline. In most cases, the disease is caused by an abnormal protein in the brain structure which becomes toxic to nerve cells. The worst scenarios come when nearing severe progression and the end phase, reaching to certain peaks of the disease. Examples for Schizophrenia might include severe delusions and derealization, meaning that potential harm to the individual may occur, while other diseases, like Parkinson’s, can affect mobility and normal day-to-day functions.
Motivation
If you looked and assembled the statistics, would you stop for a second if you realized that 50 years from now, there is a chance that 1 of every 2 people will suffer from some sort of neurodegenerative disease. Take Alzheimer’s as an example: How many of us are aware that the first patient diagnosed with Alzheimer’s was “discovered” 170 years ago, and to this day, most people hold the opinion that it is simply part of the “normal aging process”? The truth is that it is a disease and people suffering from neurocognitive decline need every type of help they can get.
So, we are still waiting for an answer. How can I, a data engineer, help with neuroscience? The answer is simple – with data! As with every area, medicine and neurology rely on the analysis of data collected from the brain’s processes – collected from pure experience. By making solutions for the management and analysis of that data, a data engineer can be a crucial puzzle piece in research as well as real-time assistance. It is important to note that many behavioral aspects in these cases are not yet explained or predictable, so ongoing research and data insights can really do something to help.
A data engineer oversees the creation of solutions for managing collected brain data to help neuroscientists and data scientists find new insights, whether that be for predicting and preventing diseases, or even finding hidden parameters that could help with finding the cure. Even if a cure is not found, we can help by improving the quality of life for an individual suffering from a certain disease. A data engineer primarily works by working through Big Data, which is precisely what medical data is.
In the following section, we will get to grips with the complexity and common problems being encountered when working with neurological medical data and look at the ways to handle them best.
Neurodata and brain metrics
Developments in the fields of Neuroscience have led to a great increase in the amounts and complexity of brain data. As in general healthcare and medical data, it is even more difficult to meet the requirements to manipulate such a complex system – our brain. Raw brain data is mostly measured in signals, which can be quantified, and as with most personal medical data, complies to strict data ethics rules. Brain activity is measured in waves, oscillating electrical voltages produced by neurons.
We can distinguish 5 basic wave types in the brain: alpha, beta, gamma, delta, and theta, which are simultaneously present depending on the type of activity a person is doing and vary based on frequency. We measure the brain using three most frequently used methods: EEG, MEG and fMRI. Electroencephalography (EEG) is a technique in which small detectors, called electrodes, are placed on a person’s scalp using a cap or a headset. This technique measures the electrical activity of groups of neurons that transmit similar electrical signals at the same time and is represented as images (signals) which can later be translated to quantitative representation (qEEG), better known as brain mapping. Magnetoencephalography (MEG) is a technique which uses the magnetic field generated by the neuron’s electrical activity and results in a more in-depth image of the brain structure, and therefore can discover deeper abnormalities, such as tumors etc. fMRI (functional Magnetic Resonance Imaging) measures brain function by detecting changes in blood flow associated with neural activity – blood flow in a certain brain region increases when that area is in use.
These brain measuring methods, although initially represented as images and signals, can be observed, and processed as numerical data. For simplicity, we will refer to an EEG example in the following sections.
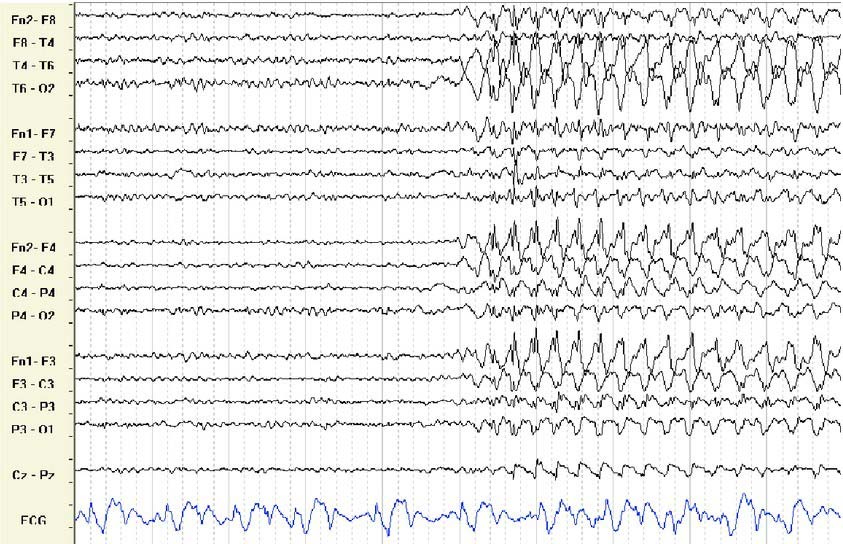
An example of raw EEG data is shown below in .edf format. It is divided into pairs of electrodes, which measure potential between different spots on the head (brain).
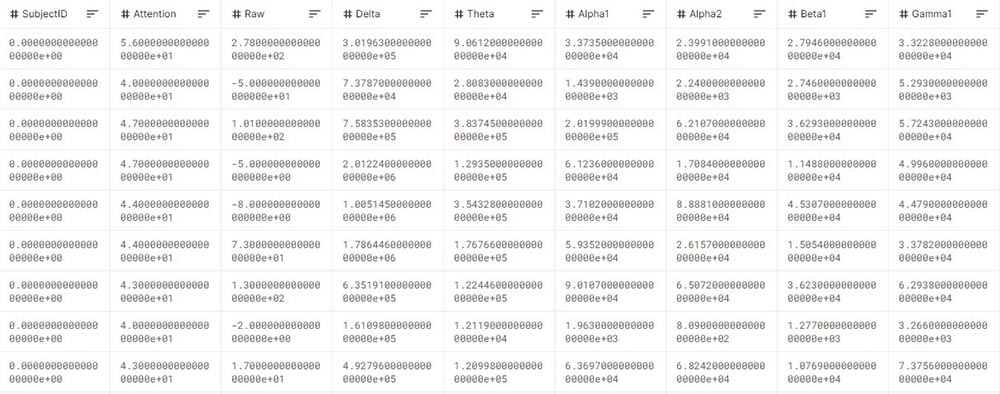
Here is an example of EEG data in a table (numeric) format. For this to be possible, the data layout should be standardized and adapted to the different EEG systems. In this case, attention represents the measure of mental focus, raw – the raw EEG signal converted to numerical form, and it is followed by presence amounts of certain brain waves, recognized in frequencies.
The EEG community uses various formats for storing raw data and there is no such standard that all researchers agree on. A common one is the aforementioned European data format, (.edf) used for storing biosignals. A collection of data related to an EEG measurement is everything but simple and concise – along with raw wave measurements, comes a range of metadata and connected datasets, needed for describing the measurement conditions and the additional information regarding it, for example, event, channels, electrodes, and system description, as well as the patient’s profile.
Here is a section of a JSON file that describes the event of running in order to measure types of brain activity, which occur during that task.
Complexity and handling
The overall data in the medical industry, especially neuro data, is, as we’ve seen in the above examples, extremely complex and can be referred to as big data. It is characterized as heterogeneous, source variant, structurally different, inter-connected and dependable on many factors. The “trigger” or an event is just one piece of information among many other factors which cause different impulses and reactions in the brain.
A single neuroimaging dataset can be measured in terabytes, so storing and handling it are challenges – but so is making it useful for “cracking” the brain’s code. There are techniques for shrinking and utilizing that data, for example isolating each neuron and assigning timestamps, but each technique creates big data, which is still very computationally demanding.
Nevertheless, the raw data described above isn’t that significant in comparison to algorithms and later processing that show the use of it.
It is clear that we have too much and too little data at the same time, to address the brain’s complexity.
The data we are handling here is unstructured, consists of images, texts, signals, streams, numerical values, metadata, and can also be referred with different scales, systems, and environments. All neuro data is large in volume and has significant variety. Imagine that if we observe the brain during an EEG, whose signals are divided into different waves dependent on different types of actions – we will have a largely scaled dataset for every case, right down to basic functions like blinking. Velocity i.e., the speed of analytical processing is also important, especially when it comes to real-time healthcare systems. Sensors produce a huge volume of, streamed data, in RT systems, that needs to be processed immediately, not with batch processing. Regarding the veracity parameter or “the uncertainty” of the data – large data collections are often combined from various and sometimes unreliable sources, therefore making its proper analysis even more difficult.
These four aspects of neuro data are, in fact, aspects of big data, therefore we can and need to apply big data methods accordingly.
Things like an individual’s position or feelings may be irrelevant to other studies, but in cases of neurology cannot be neglected. Every piece of information could be helpful for recognizing hidden patterns, so it is a must to properly manage and exploit the variety of this data. Data collected from the brain for neuronal models rely both on experimental and computational methods and because of that – simulation and data modeling have a significant role. Following that fact, data sharing in the neuroscientific community is essential for data-driven approaches to neuroscience.
Since this is heterogeneous data, used in different environments and aspects, it is important to utilize it across different environments and standardize data models, as well as preserve the dedication to constantly levelling up our available knowledge base. Medicine generates terabytes of data, and proper management and organization of information is mandatory to preserve that state and achieve future discoveries.
Data Ethics
Neuroethics is a field that studies the ethical, legal, and societal implications of neuroscience, and observes ways in which neurotechnology can be used to predict or alter human behavior. Some neuroethical problems are not fundamentally different from those encountered in bioethics
– morals in medical policy and practice. Others are unique to neuroethics because the brain, as the organ of the mind, has implications for broader philosophical problems, such as the nature of free will, moral responsibility, self-deception, and personal identity, which need to be addressed in real-time. Neuroscience gets to the questions of the nature of living, mental privacy, identity features and others whose characteristics seemed non-changeable, but nowadays are harder to define. There is a need to engage in data sharing for neuroscience, but we need to respect certain standards, basic values, and cultures, because gaps in understanding can lead to fewer or missed opportunities and neuroscientific findings. An example of goals would include the identification of values and principles or strategizing what values to observe that could change data governance and insights.
We can look back on some of the ethical insufficiencies of the current laws as well as difficulties in ethical tool development, which include questions that haven’t yet been addressed like: How to respect an individual and their values? What is an acceptable data processing practice? Does neuroscience data need special practices? What kinds of data raise ethical concern? How to clarify ethical issues throughout the data lifecycle? How to create practical, ethical, and universal tools for this data?
Along with data sharing and governance, data ethics comes into the equation when creating a good data-driven model of the brain.
DATA-DRIVEN MODEL
Modeling a human brain is very challenging, because the data is non-stationary and dependent on various parameters, as well as limited to ethical standards. The development of data-driven models has the potential to transform neural engineering applications to rehabilitate, assist, and even upgrade the human nervous system. Most of the methodologies for understanding and efficiently using neural data rely on its quantitative understanding – for example, complex brain EEG signals are explained as numeric differences of potential between different locations in the brain. The data driven model of the brain, which is a representation of reality, should basically be a representation of data held in the brain, so the choice of a basic model vastly depends on the amount of recorded data, as well as amount of knowledge available in that aspect.
Brain models can have different levels – ranging from molecular, synapse, circuit, and system levels. Here we will describe an example of a data model of a brain, stating the elements it should consist of. The model should have high interoperability and should be observer independent. The data held in it should be of many different data types and highly representative and described through metadata. For the model to be implementable and applicable in different neural communities (for example in research and clinical), where different methods can be used, we will need to provide sets of data descriptors, such as relations between data (object-relational model). It is a must to implement relations like “extends”, so the data can be referenced outside a single database onto different databases and registries. The data model includes knowledge representation only secondarily because it is needed for data specification. This excludes ontological definitions as well as is-a and has-a relationships, for example: synapse is-a relation between two neurons. The design decision recognizes that the neuroscientific knowledge base is highly complex, dependent on interactions among multiple levels of anatomic organization and functional systems.
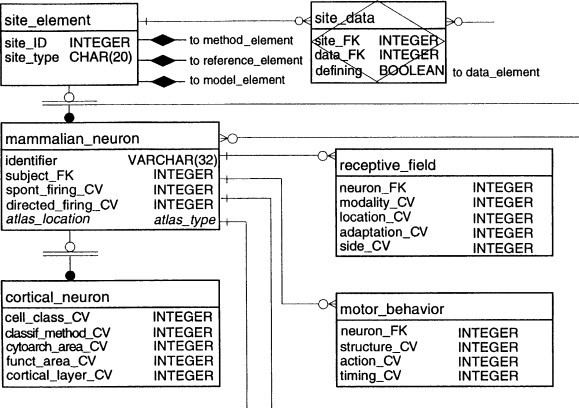
Below we can see an entity-relationship diagram of a somatosensory database. Site element abstracts neuronal anatomy. Subclasses of site elements are shown with names and data types of characteristic data and descriptive metadata attributes.
Interoperability in neuroscience data comes in handy for providing universal standards in data exchange, as well as describing universal data models, formats, and various contexts of different resources. The methods used should allow easy identification of the data type, source, and context for interpretation, without requiring changes in the data model. Such standards should permit dataset exchange and querying, while holding up consistency. Both standards and individual focused schemas should be describable in the meta-representation and expressible via interfaces. Data must be structurally organized – it can be structured in either a “flat file,” a tabular formation, a structured file (such as XML), an object based, or a layer-oriented scheme.
A sample interface model for neuro data is characterized as: extensible – meaning it is not limited to one domain of neuroscience, compact – containing data descriptors, is efficient, simple, and scalable, uses limited time and space resources and is useful for small and large projects, with a platform independent data exchange standard and convertible formats, and in the end – intuitive, familiar, and human-readable.
A neuro-database is an extremely complex relational database and is mostly referred to as a specialized database, contrary to primary and secondary databases, and therefore is held by special labs or consortiums. Such databases are characterized with a research-specific relational schema and specialized data types. They are under constant development making it a challenge for designers that must support data querying, acquiring, and parsing from established data sources, as well as for integrating the results with their own data model.
A possible framework example should include structuring experimental data with standardized models and incorporating the experimental data and models with other relevant data (diseases, articles, and biological models). In the context of neuroinformatics, we can take various types of experimental data, encapsulate it into interconnected classes (creating a schema), and then link each of them to a structured data model (like the one described above).
DATA ENGINEERING
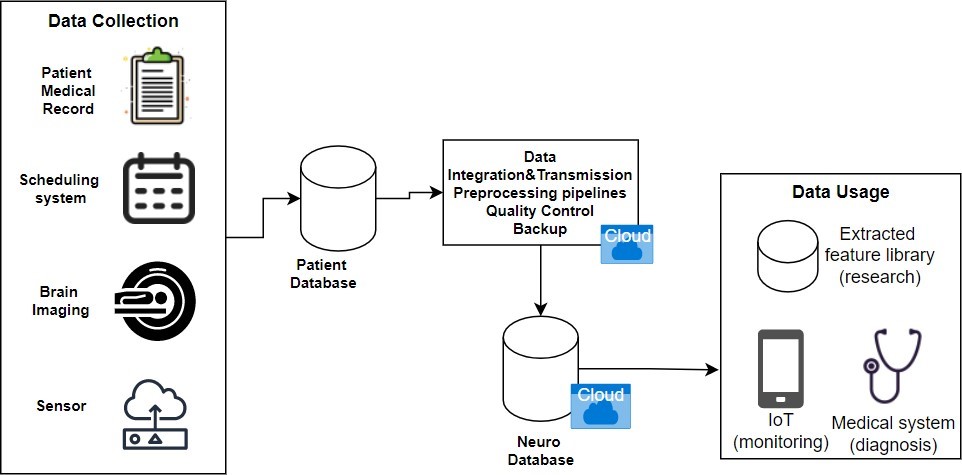
Coming from a need for data insight in all neurology aspects, from collecting and processing medical data to big data governance, the hand of a data engineer in neurology is inevitable. Since neuro data is large in scale and highly complex, proper handling of it, for various uses, is a priority. In terms of data engineering, we can discuss data collecting, data modeling, data preprocessing, data ethics, and other processes. Use cases range in biomedical informatics systems, IoT and embedded systems (real-time monitoring), medical technology (medical devices), data governance for large systems and preprocessing for machine learning models. All of them come into the equation with data engineering.
Data collecting and formatting
Understanding the brain requires a broad range of approaches and methods from the domains of biology, psychology, chemistry, physics, and mathematics, and the main challenge is to differentiate complex behaviors and functions, like thoughts, memories, actions, and emotions. This demands the acquisition and integration of vast amounts of data of many types, at multiple scales in time and in space.
Data mining was introduced as a non-trivial process to identify new and useful patterns in large data sets. In the case of neurology, data mining may be used for diagnosis and prognosis, therapy planning and rehabilitation, as well as image and signal analysis. Today, application in the analyses of datasets with neurological relevance is still limited and mainly focuses on the use of data mining in clinical medicine – for the identification of relations, patterns and models supporting predictions and the clinician’s decision-making processes, e.g., for diagnosis, prognosis, and treatment planning. When validated, these predictive models could be embedded in the clinical information systems as clinical decision support modules.
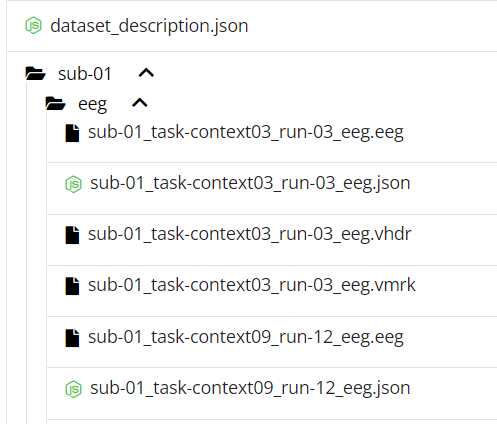
Another problem in neuroscience industry is handling data collected in various and omnipresent formats described in the sections above. There is no consensus how to organize data obtained from neuroimaging methods and the lack of it leads to various misunderstandings on how to arrange and observe data. Looking at the example “triplet” data of a single brain recording which is consisted of a binary .eeg file (EEG recordings, voltage values) and text .vhdr (metadata like amplifiers, filters, channel number..) and .vmrk (markers for eeg data, event information) files, it is clear that raw brain metrics aren’t something which is easy to understand and observe by an average user. This given example is contained in the BIDS (Brain Imaging Data Structure) format shown in the picture below, which describes a simple way of organizing neuroimaging data.
We can say that the two main challenges are using these inconsistent data formats against universal formats, like csv or JSON, and respecting the relationships between these files. An entity-linked file collection is a good solution for organizing relationships between these files. These collections are identified with a suffix indicating the group of files that need to be considered a logical unit. The first challenge remains, since brain recordings cannot be just “translated” to a widely used format and various tools and algorithms are required to process that data. Generally, the data obtained from different manufacturers often requires various scripts to convert from the manufacturers format into the BIDS format, which is mostly used.
Data governance and Biomedical Informatics
The primary goal of data governance is to maintain the quality and usability of data and biomedical informatics benefits from it in the medical industry to provide clinical insights, uncover disease, treatment and response patterns and point to new lines of scientific and medical inquiry. It is scary on one hand, that today’s hospitals would probably crash if it wasn’t for the skilled maintenance of their computer systems and databases. Biomedical informatics therefore includes big data, generating and managing terabytes of it, as well as constant system maintenance in all types of medical industries. Also, exploiting big data technologies, large scale databases and parallel computing, are as necessary for large research centers as they are for everyday hospitals. Biomedical Informatics primarily take care of basic clinical data – patient’s records (EMR – Electronic Medical Record), statuses and device-dependent functions.
Integrated bioinformatics on the other hand, focuses on solving data integration problems for life sciences, for example – developing tools for automated literature and diagnostical analysis. That area is a “big data” analysis area, in which volume and variety play an essential role. All neuro data has sources from government funded labs, with elaborated data to individual small labs, with limited data sets that include unpublished “dark-data”. As neuroscience data insights need to be shared, deepening this data (yielding useful information from it) is extremely hard but necessary.
Big Data Technologies, Software, Algorithms and Architectures
To exploit neuro data for its full potential, there is a need to use and develop a wide range of technologies and algorithms to help with that task. Here we will cover just some of them, like Map-Reduce, Hadoop, NoSQL databases, and cloud computing, which are likely to be used to improve the performance of healthcare IT systems and research processes.
A potentially successful way to handle this data would be Map-Reduce, a programming model aiming at simplifying parallelization. By organizing computational steps using two functions map (mapping key-value pairs) and reduce (processing pairs with the same key to derive the final result), the algorithm is simply implemented and moves away from the need to use fine tuning of low-level programming languages in parallel programming, which is needed in neuroscience. That said, Map Reduce still requires a complex architecture, with a core distributed file system.
The Apache software foundation has developed one of the most widely used Map-Reduce implementations: Hadoop. An example use case of map-reduce method is simplifying computational steps in cognitive disease prediction, like Alzheimer’s, which heavily relies on statistics and complex equations by parallelizing computation steps.
New database technologies are also providing solutions to the scalability problem. For example, NoSQL data management systems are designed to provide easy horizontal scaling to represent data without relational modeling. The usage of relational databases has its disadvantages when storing heterogeneous data. This is solved by using document-oriented databases, like MongoDB.
In today’s neuroscience data research, there are two models – the traditional and the cloud model. In the traditional one, data is generated by one group of researchers and shared with the others, where each group develops its own tools for data analysis and governance and data is often stored in local repositories. In the cloud-based model, the infrastructure is switched from local repositories to cloud, which expands access to data and attracts more researchers. In this case they use a common set of tools and work with data located in a platform. Neuro data is stored in cloud by healthcare institutions, laboratories and even hospitals, while respecting patient consent and general sharing restrictions – for example, highly restricted databases are ones holding data of neurocognitive disease cases.
Together with software and data management tools, new architectures are also necessary to support big data management and analysis; in particular, cloud-computing seems a crucial solution to enable high performances while containing building and operational costs. Currently, there are several cloud types, which may be suited for almost all needs in healthcare and biomedicine. Cloud environments for healthcare along with cloud computing may surely provide an excellent, connected, and accessible environment for patients in need, as well as for doctors and researchers.
Many of the challenges healthcare companies and research centers face, due to high complexity data management, are high cost, storage demand, accessibility and security. Tons of data, collected from devices, that needs to be stored and processed, requires high hardware demands, so providers like Azure and AWS offer many cloud services that cost as much as healthcare uses them. However, due to data stream being constant and large in scale, this isn’t necessarily a good thing. Regardless, thanks to cloud services, data can be updated and collected in real-time (e.g. Azure IoT Hub), and every researcher and specialist can access it.
There is always a risk of breakdowns and data loss with a no restore option in the case of hardware resources – this can cause irreparable consequences. In the case of cloud services, the provider offers backups through servers.
Therefore, most of these listed big data handling requirements, such as computing, analytics, security and storage options, are covered with cloud providers. Today, AWS is the most trusted platform for healthcare providers and protected health information. It offers help with clinical systems (Electronic Health Records), by including NLP and machine learning to optimize administrative tasks and medical imaging, by migrating it to cloud and optimizing data extraction. Some the services used here are Amazon S3 as a storage solution, Amazon EC2 as a computing service and the recently developed Amazon HealthLake for transforming, querying and analyzing health data.
Although cloud is a good solution, it still has its cons and legal, ethical, and administrative challenges. This kind of data cannot be easily shared, and a requirement that different frameworks and models are needed to protect patient’s privacy. To analyze data the tools are brought to the data rather than permitting that the data can be downloaded. A large amount of complex neuroimaging data requires quality analytical techniques in cloud use. As a result, several cloud-based infrastructures have been developed for the management of neuroimaging data. For example, there are some developed cloud based computation environments for neuroimaging data processing and visualization. And lastly – the economics of cloud-based computing for neuroimaging remains a question, comparing and concluding if high-cost cloud computing is better than computer infrastructure held in a lab.
The cloud model has led to a vast increase in the quantity and complexity of data and expanded access to it, which has attracted many more researchers, enabled multi-national neuroscience collaborations, and induced the development of many new tools.
Technology in medicine – medical devices and IoT
With the latest microcontrollers and embedded processors, medical device manufacturers have an opportunity to improve patient health care and the productivity of clinicians. Devices used in neurology include neurodiagnostic, neurointerventional and neurostimulation devices, ranging from “simple” monitoring devices (wearable monitors) to complex diagnostic machines, like magnetoencephalograms.
Still, data management is a must for every mission-critical device, which demands safe, reliable, scalable storage, and real-time response. Today, with the growing complexity and “ambition” of medical devices, they now focus on making decisions
autonomously using accumulated data about one or more patients. To accomplish this while ensuring patient safety and privacy, it is important to select an appropriate database technology, and have access to relevant knowledge. A data management solution is required, due to specific requirements such as: frequently updated and revised patient information, collected data exceeding available memory, safe recoveries during failures, concurrent tasks reading and writing to a collection of records, filtering and analyzing medical records through graphs and reports, and continuously distributed data for system and device synchronization. An embedded database management system integrates directly with the firmware on a medical device to store and access data without an administrative component. In fact, it is similar to a file system, which stores data in a flat or hierarchical format, such as CSV, XML, or a custom binary format.
IoT today comes to the forefront in the diagnostic and real-time monitoring industry. By collecting information through sensors, it can help in standard brain measurement techniques as well as seizure prediction, creating a wide range of algorithms that help researchers and doctors, and improve the overall quality of an individual’s life. Of course, nothing would be possible without the proper real-time data management described above. Cloud providers also hold IoT solutions, like Azure’s IoT Hub which offers methods to capture and analyze brain waves. To assist people with neurocognitive disabilities, or even the ones with normal functioning, the Brain-Computer interfaces (BCI) come to mind. By acquiring brain signals and translating them into commands for connected devices, such implementations can really make brain-related wonders which we were not even aware of.
Conclusion
By applying engineering principles and knowledge, we as data engineers can be of help in Neuroscience, a very important, vital industry, which is “forced” to work with not just big data, but extremely complex and demanding data. Along with accepting the cultural shift – the need for data sharing and data acquiring, raising awareness about its importance, and contributing to the processing, managing and development of engineering techniques, tools and architectures which benefit cases in neurology, a data engineer can be crucial to future research progress and clinical application within neuroscience and neurology.
170 years have passed with little progress in curing a disease like Alzheimer’s, and maybe the same amount of time will pass again without a solution, but our involvement in research studies and healthcare processes can make a small and significant step into helping the brain,and recognizing a potential solution for neurodegenerative cases.